Autochthonous Plasmodium vivax Infections, Florida, USA, 2023
Azhar Muneer
1, Swamy R. Adapa
1, Suzane Silbert, Kelly Scanlan, Harold Vore, Andrew Cannons, Andrea M. Morrison, Danielle Stanek, Carina Blackmore, John H. Adams, Kami Kim
2, Rays H.Y. Jiang
2, and Liwang Cui
2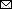
Author affiliations: University of South Florida Morsani College of Medicine, Tampa, Florida, USA (A. Muneer, K. Kim, L. Cui); University of South Florida School of Public Health, Tampa (S.R. Adapa, J.H. Adams, K. Kim, R.H.Y. Jiang); Tampa General Hospital, Tampa (S. Silbert, K. Kim); Sarasota Memorial Hospital, Sarasota, Florida, USA (K. Scanlan, H. Vore); Florida Department of Health Bureau of Public Health Laboratories, Tampa (A. Cannons); Florida Department of Health, Tallahassee, Florida, USA (A.M. Morrison, D. Stanek, C. Blackmore)
Main Article
Figure 2

Figure 2. Phylogenetic analysis of Plasmodium vivax strains from blood samples from malaria patients, Florida, USA, May–July 2023, suggesting Central/South America origin. A) Geographic distribution of 53 high-quality global strains selected from >1,000 global P. vivax collections. B) Florida P. vivax strains clustering with Central/South America strains. The phylogenetic tree was constructed by using the maximum-likelihood method, and 1,000 bootstrap replications are shown next to the branches. The color coding of the geographic origin of the isolates matches the global map in panel A. The US and Central/South American cluster is shaded gray. The 4 Florida P. vivax strains are denoted as 1AS1, 2AS2, 3AS3, and 4AS4. Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: April 25, 2024
Page updated: May 22, 2024
Page reviewed: May 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
